Iron »
PDB 9ina-9kpp »
9iss »
Iron in PDB 9iss: Crystal Structure of Cytochrome P450BM3 III-10C1 Mutant Heme Domain with N-Decanoyl-L-Homoserine Lactone
Enzymatic activity of Crystal Structure of Cytochrome P450BM3 III-10C1 Mutant Heme Domain with N-Decanoyl-L-Homoserine Lactone
All present enzymatic activity of Crystal Structure of Cytochrome P450BM3 III-10C1 Mutant Heme Domain with N-Decanoyl-L-Homoserine Lactone:
1.14.14.1; 1.6.2.4;
1.14.14.1; 1.6.2.4;
Protein crystallography data
The structure of Crystal Structure of Cytochrome P450BM3 III-10C1 Mutant Heme Domain with N-Decanoyl-L-Homoserine Lactone, PDB code: 9iss
was solved by
Y.Yokoyama,
O.Shoji,
H.Sugimoto,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 47.44 / 1.46 |
| Space group | P 1 21 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 58.575, 145.442, 63.106, 90, 97.37, 90 |
| R / Rfree (%) | 16.6 / 18.2 |
Iron Binding Sites:
The binding sites of Iron atom in the Crystal Structure of Cytochrome P450BM3 III-10C1 Mutant Heme Domain with N-Decanoyl-L-Homoserine Lactone
(pdb code 9iss). This binding sites where shown within
5.0 Angstroms radius around Iron atom.
In total 2 binding sites of Iron where determined in the Crystal Structure of Cytochrome P450BM3 III-10C1 Mutant Heme Domain with N-Decanoyl-L-Homoserine Lactone, PDB code: 9iss:
Jump to Iron binding site number: 1; 2;
In total 2 binding sites of Iron where determined in the Crystal Structure of Cytochrome P450BM3 III-10C1 Mutant Heme Domain with N-Decanoyl-L-Homoserine Lactone, PDB code: 9iss:
Jump to Iron binding site number: 1; 2;
Iron binding site 1 out of 2 in 9iss
Go back to
Iron binding site 1 out
of 2 in the Crystal Structure of Cytochrome P450BM3 III-10C1 Mutant Heme Domain with N-Decanoyl-L-Homoserine Lactone
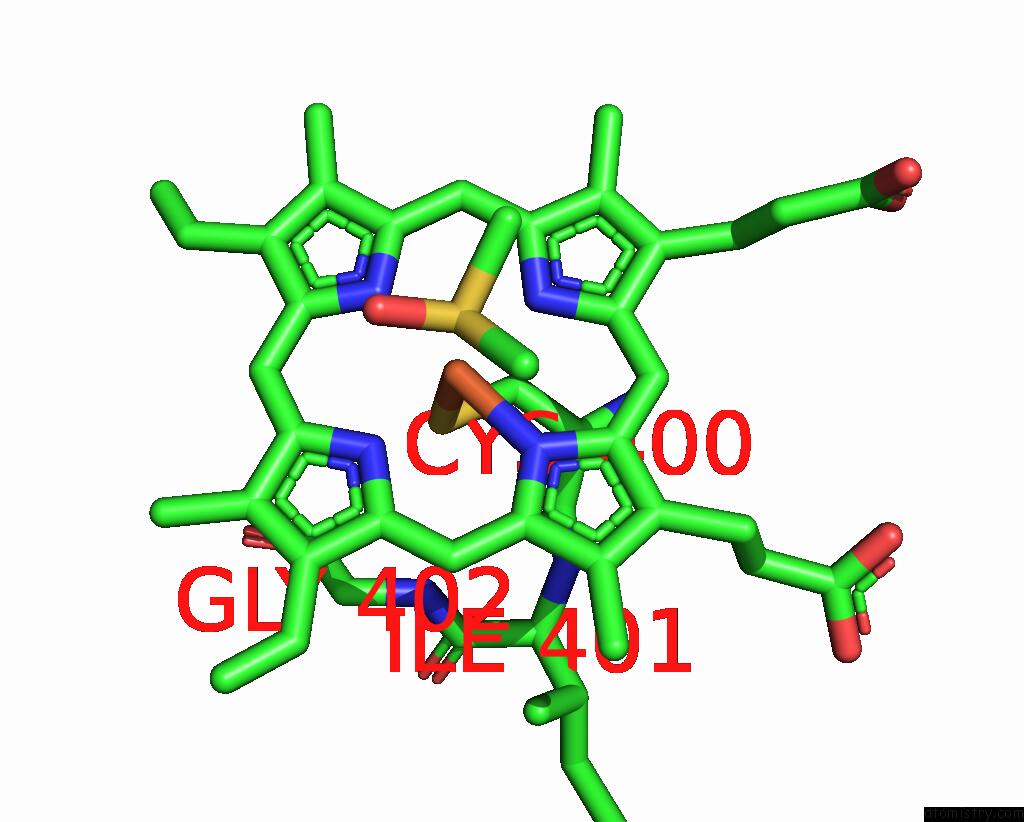
Mono view
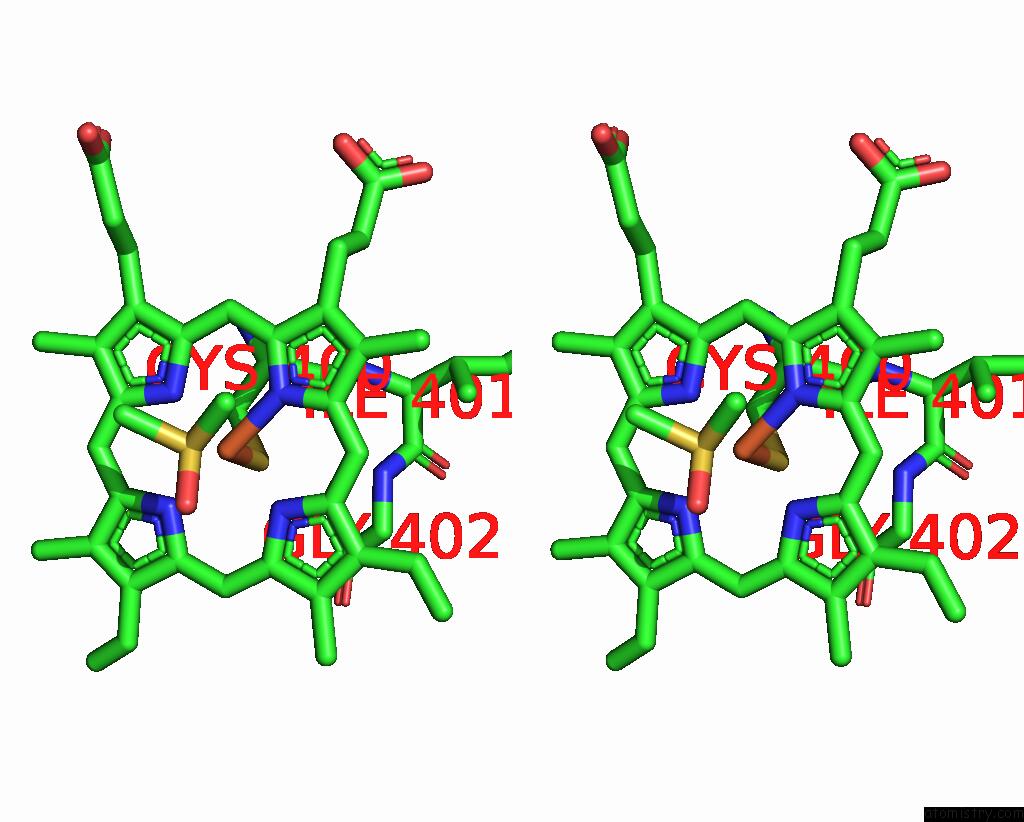
Stereo pair view

Mono view

Stereo pair view
A full contact list of Iron with other atoms in the Fe binding
site number 1 of Crystal Structure of Cytochrome P450BM3 III-10C1 Mutant Heme Domain with N-Decanoyl-L-Homoserine Lactone within 5.0Å range:
|
Iron binding site 2 out of 2 in 9iss
Go back to
Iron binding site 2 out
of 2 in the Crystal Structure of Cytochrome P450BM3 III-10C1 Mutant Heme Domain with N-Decanoyl-L-Homoserine Lactone
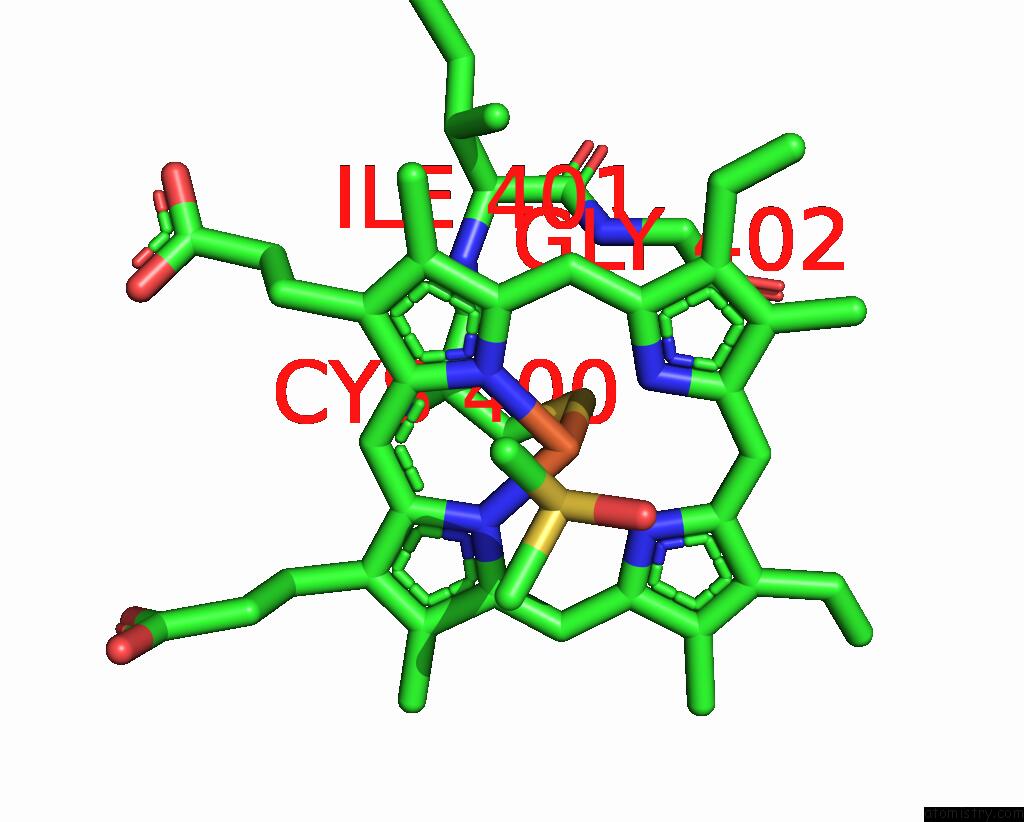
Mono view
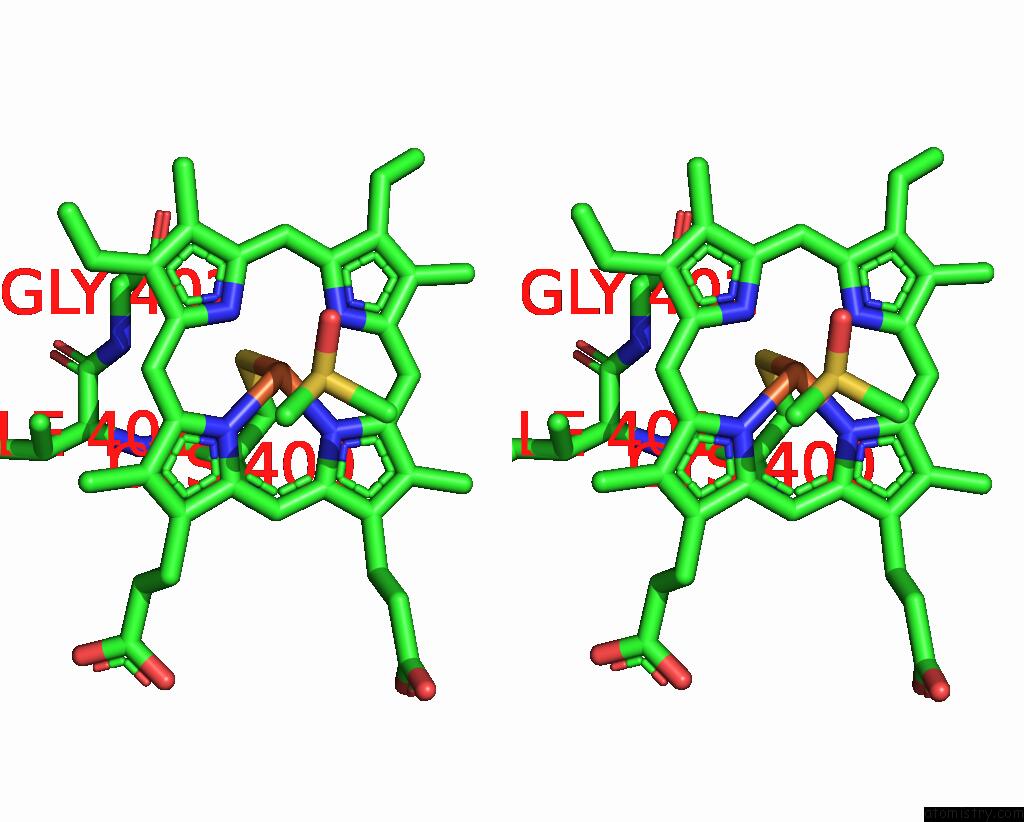
Stereo pair view

Mono view

Stereo pair view
A full contact list of Iron with other atoms in the Fe binding
site number 2 of Crystal Structure of Cytochrome P450BM3 III-10C1 Mutant Heme Domain with N-Decanoyl-L-Homoserine Lactone within 5.0Å range:
|
Reference:
Y.Yokoyama,
S.Ariyasu,
M.Karasawa,
C.Kasai,
Y.Aiba,
H.Sugimoto,
O.Shoji.
Bacterial Acyl Homoserine Lactones Triggered Non-Native Substrate Hydroxylation Catalyzed By Directed-Evolution-Derived Cytochrome P450BM3 Mutants Chemcatchem 2024.
ISSN: ESSN 1867-3899
DOI: 10.1002/CCTC.202401641
Page generated: Fri Aug 8 06:54:35 2025
ISSN: ESSN 1867-3899
DOI: 10.1002/CCTC.202401641
Last articles
I in 4BSWI in 4AW7
I in 4BSV
I in 4B43
I in 4B9H
I in 4AS2
I in 4AS5
I in 4AX2
I in 4ARR
I in 4AQ3