Iron »
PDB 1dxt-1ea1 »
1e8e »
Iron in PDB 1e8e: Solution Structure of Methylophilus Methylotrophus Cytochrome C''. Insights Into the Structural Basis of Haem-Ligand Detachment
Iron Binding Sites:
The binding sites of Iron atom in the Solution Structure of Methylophilus Methylotrophus Cytochrome C''. Insights Into the Structural Basis of Haem-Ligand Detachment
(pdb code 1e8e). This binding sites where shown within
5.0 Angstroms radius around Iron atom.
In total only one binding site of Iron was determined in the Solution Structure of Methylophilus Methylotrophus Cytochrome C''. Insights Into the Structural Basis of Haem-Ligand Detachment, PDB code: 1e8e:
In total only one binding site of Iron was determined in the Solution Structure of Methylophilus Methylotrophus Cytochrome C''. Insights Into the Structural Basis of Haem-Ligand Detachment, PDB code: 1e8e:
Iron binding site 1 out of 1 in 1e8e
Go back to
Iron binding site 1 out
of 1 in the Solution Structure of Methylophilus Methylotrophus Cytochrome C''. Insights Into the Structural Basis of Haem-Ligand Detachment
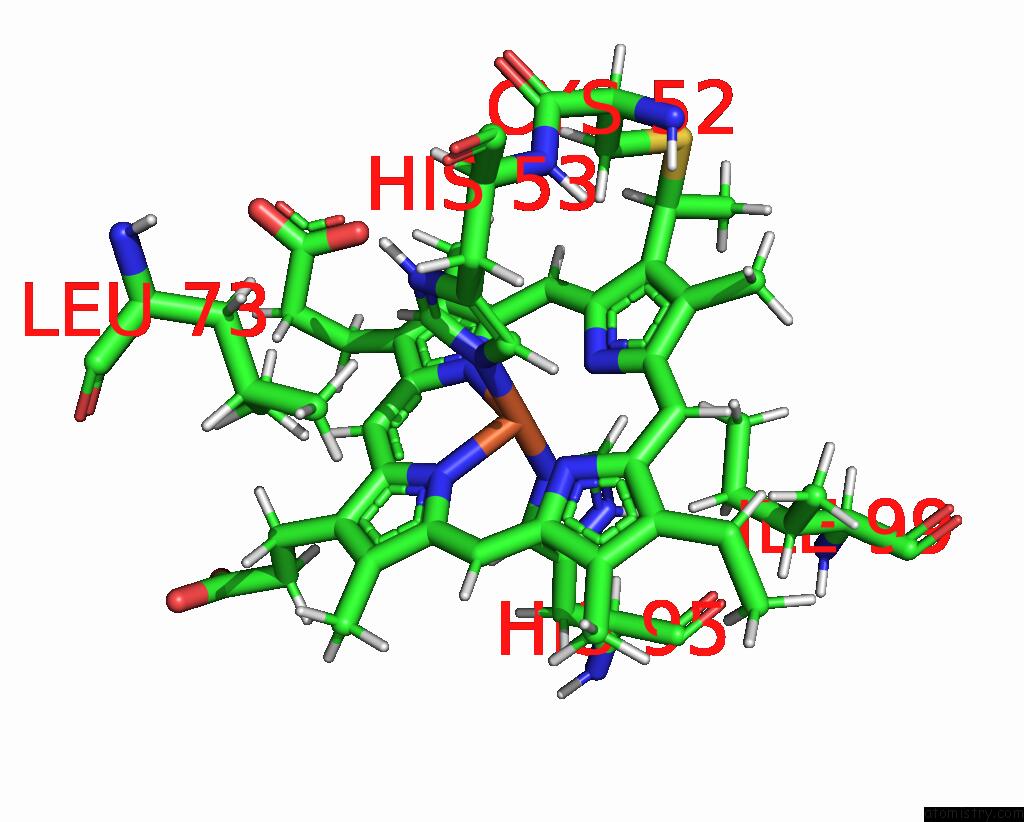
Mono view
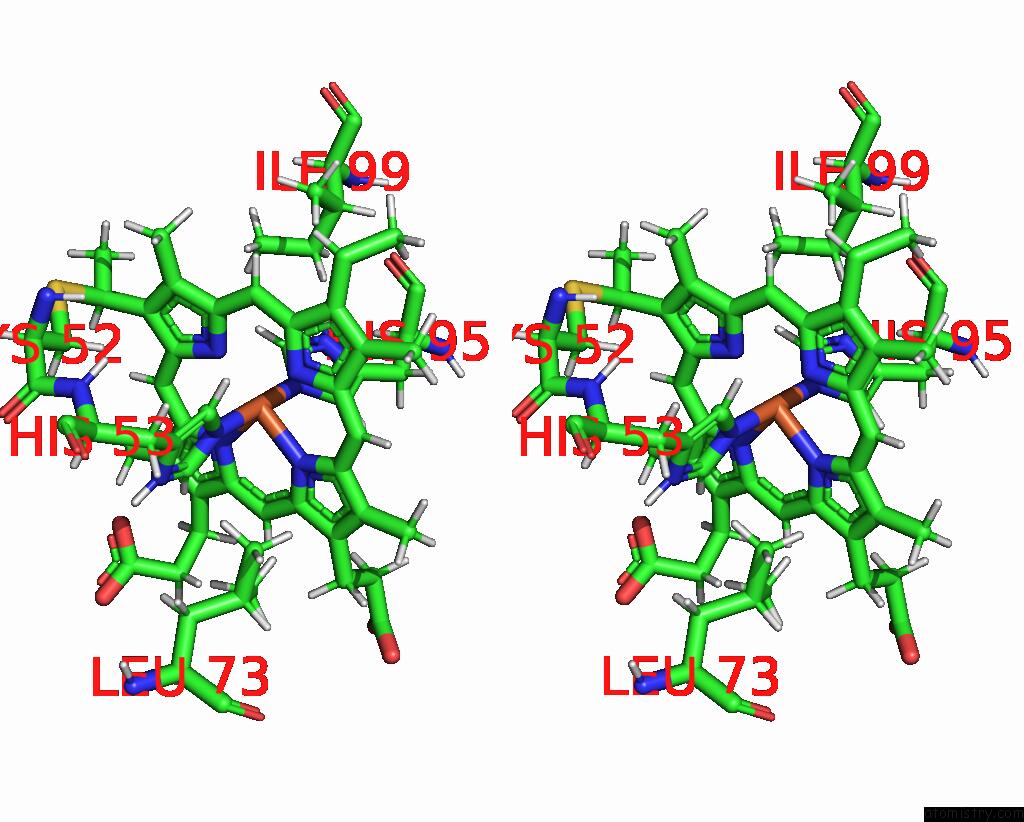
Stereo pair view

Mono view

Stereo pair view
A full contact list of Iron with other atoms in the Fe binding
site number 1 of Solution Structure of Methylophilus Methylotrophus Cytochrome C''. Insights Into the Structural Basis of Haem-Ligand Detachment within 5.0Å range:
|
Reference:
L.Brennan,
D.L.Turner,
P.Fareleira,
H.Santos.
Solution Structure of Methylophilus Methylotrophus Cytochrome C": Insights Into the Structural Basis of Haem-Ligand Detachment J.Mol.Biol. V. 308 353 2001.
ISSN: ISSN 0022-2836
PubMed: 11327772
DOI: 10.1006/JMBI.2001.4600
Page generated: Wed Jul 16 13:40:09 2025
ISSN: ISSN 0022-2836
PubMed: 11327772
DOI: 10.1006/JMBI.2001.4600
Last articles
Fe in 1QKCFe in 1QJS
Fe in 1QJQ
Fe in 1QJF
Fe in 1QJE
Fe in 1QJD
Fe in 1QIQ
Fe in 1QI8
Fe in 1QHW
Fe in 1QHU