Iron »
PDB 6cxv-6dhz »
6d7k »
Iron in PDB 6d7k: Complex Structure of Methane Monooxygenase Hydroxylase in Complex with Inhibitory Subunit
Protein crystallography data
The structure of Complex Structure of Methane Monooxygenase Hydroxylase in Complex with Inhibitory Subunit, PDB code: 6d7k
was solved by
H.Kim,
S.J.Lee,
U.-S.Cho,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 22.29 / 2.60 |
| Space group | C 1 2 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 179.781, 125.776, 126.380, 90.00, 102.89, 90.00 |
| R / Rfree (%) | 17.8 / 22.1 |
Iron Binding Sites:
The binding sites of Iron atom in the Complex Structure of Methane Monooxygenase Hydroxylase in Complex with Inhibitory Subunit
(pdb code 6d7k). This binding sites where shown within
5.0 Angstroms radius around Iron atom.
In total 4 binding sites of Iron where determined in the Complex Structure of Methane Monooxygenase Hydroxylase in Complex with Inhibitory Subunit, PDB code: 6d7k:
Jump to Iron binding site number: 1; 2; 3; 4;
In total 4 binding sites of Iron where determined in the Complex Structure of Methane Monooxygenase Hydroxylase in Complex with Inhibitory Subunit, PDB code: 6d7k:
Jump to Iron binding site number: 1; 2; 3; 4;
Iron binding site 1 out of 4 in 6d7k
Go back to
Iron binding site 1 out
of 4 in the Complex Structure of Methane Monooxygenase Hydroxylase in Complex with Inhibitory Subunit
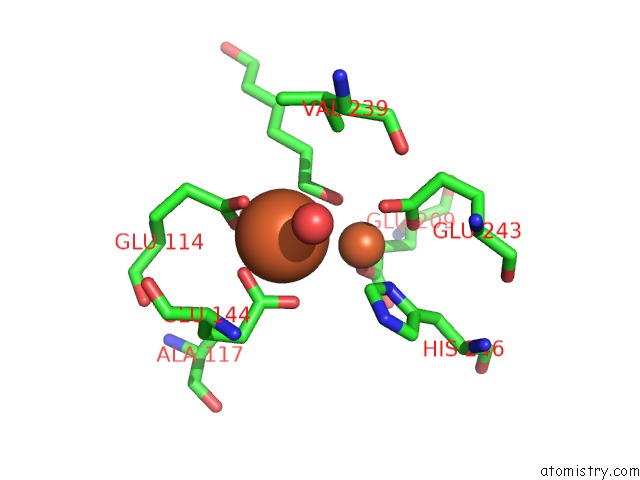
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Iron with other atoms in the Fe binding
site number 1 of Complex Structure of Methane Monooxygenase Hydroxylase in Complex with Inhibitory Subunit within 5.0Å range:
|
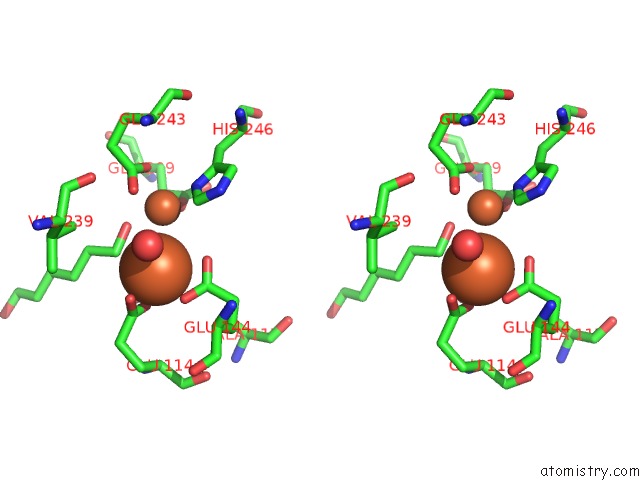
Iron binding site 2 out of 4 in 6d7k
Go back to
Iron binding site 2 out
of 4 in the Complex Structure of Methane Monooxygenase Hydroxylase in Complex with Inhibitory Subunit

Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Iron with other atoms in the Fe binding
site number 2 of Complex Structure of Methane Monooxygenase Hydroxylase in Complex with Inhibitory Subunit within 5.0Å range:
|
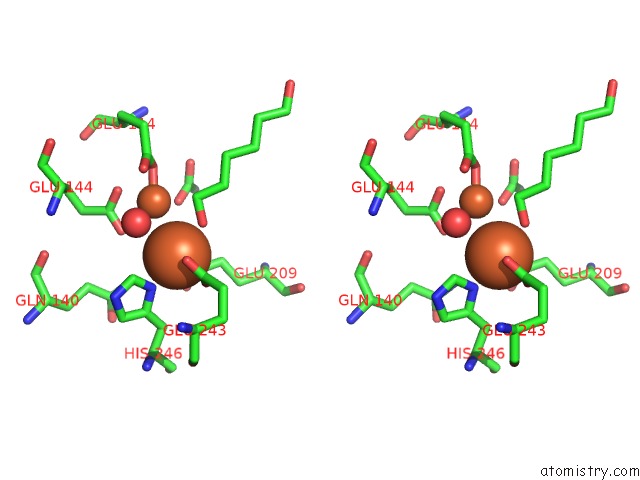
Iron binding site 3 out of 4 in 6d7k
Go back to
Iron binding site 3 out
of 4 in the Complex Structure of Methane Monooxygenase Hydroxylase in Complex with Inhibitory Subunit

Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Iron with other atoms in the Fe binding
site number 3 of Complex Structure of Methane Monooxygenase Hydroxylase in Complex with Inhibitory Subunit within 5.0Å range:
|
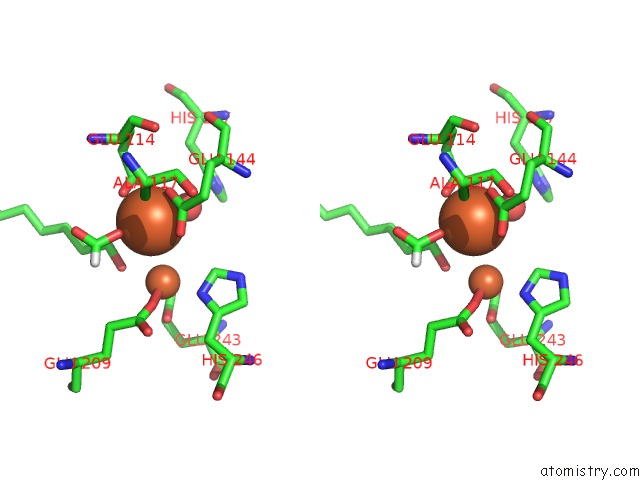
Iron binding site 4 out of 4 in 6d7k
Go back to
Iron binding site 4 out
of 4 in the Complex Structure of Methane Monooxygenase Hydroxylase in Complex with Inhibitory Subunit

Mono view
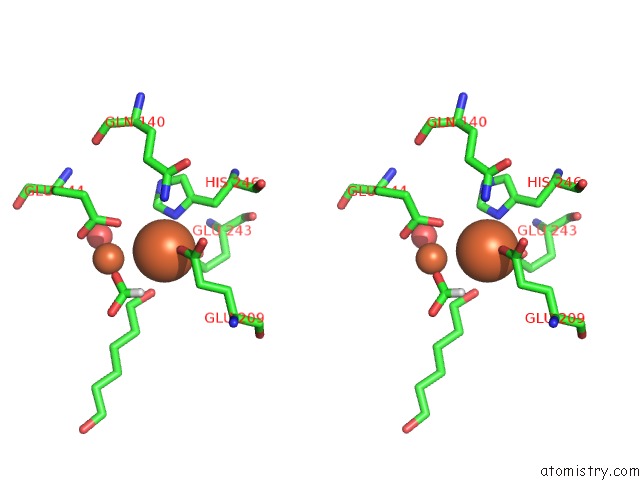
Stereo pair view

Mono view

Stereo pair view
A full contact list of Iron with other atoms in the Fe binding
site number 4 of Complex Structure of Methane Monooxygenase Hydroxylase in Complex with Inhibitory Subunit within 5.0Å range:
|
Reference:
H.Kim,
S.An,
Y.R.Park,
H.Jang,
H.Yoo,
S.H.Park,
S.J.Lee,
U.S.Cho.
Mmod-Induced Structural Changes of Hydroxylase in Soluble Methane Monooxygenase. Sci Adv V. 5 X0059 2019.
ISSN: ESSN 2375-2548
PubMed: 31616787
DOI: 10.1126/SCIADV.AAX0059
Page generated: Wed Aug 6 05:19:12 2025
ISSN: ESSN 2375-2548
PubMed: 31616787
DOI: 10.1126/SCIADV.AAX0059
Last articles
Fe in 7DDQFe in 7D6V
Fe in 7DAO
Fe in 7D9S
Fe in 7D9R
Fe in 7D9J
Fe in 7D9I
Fe in 7D9H
Fe in 7D9G
Fe in 7D9F