Iron »
PDB 6e45-6f0a »
6ex0 »
Iron in PDB 6ex0: Crystal Structure of Relp (SAS2) From Staphylococcus Aureus Bound to Pppgpp in the Post-Catalytic State
Enzymatic activity of Crystal Structure of Relp (SAS2) From Staphylococcus Aureus Bound to Pppgpp in the Post-Catalytic State
All present enzymatic activity of Crystal Structure of Relp (SAS2) From Staphylococcus Aureus Bound to Pppgpp in the Post-Catalytic State:
2.7.6.5;
2.7.6.5;
Protein crystallography data
The structure of Crystal Structure of Relp (SAS2) From Staphylococcus Aureus Bound to Pppgpp in the Post-Catalytic State, PDB code: 6ex0
was solved by
M.C.Manav,
D.E.Brodersen,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 53.70 / 2.78 |
| Space group | I 41 2 2 |
| Cell size a, b, c (Å), α, β, γ (°) | 124.330, 124.330, 213.150, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 21.8 / 26.3 |
Iron Binding Sites:
The binding sites of Iron atom in the Crystal Structure of Relp (SAS2) From Staphylococcus Aureus Bound to Pppgpp in the Post-Catalytic State
(pdb code 6ex0). This binding sites where shown within
5.0 Angstroms radius around Iron atom.
In total only one binding site of Iron was determined in the Crystal Structure of Relp (SAS2) From Staphylococcus Aureus Bound to Pppgpp in the Post-Catalytic State, PDB code: 6ex0:
In total only one binding site of Iron was determined in the Crystal Structure of Relp (SAS2) From Staphylococcus Aureus Bound to Pppgpp in the Post-Catalytic State, PDB code: 6ex0:
Iron binding site 1 out of 1 in 6ex0
Go back to
Iron binding site 1 out
of 1 in the Crystal Structure of Relp (SAS2) From Staphylococcus Aureus Bound to Pppgpp in the Post-Catalytic State
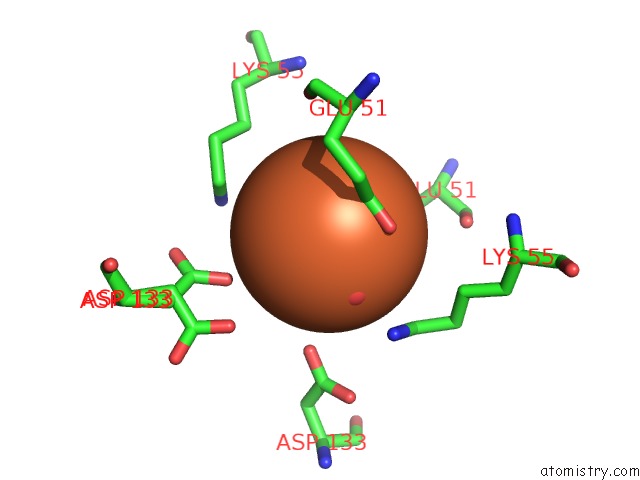
Mono view
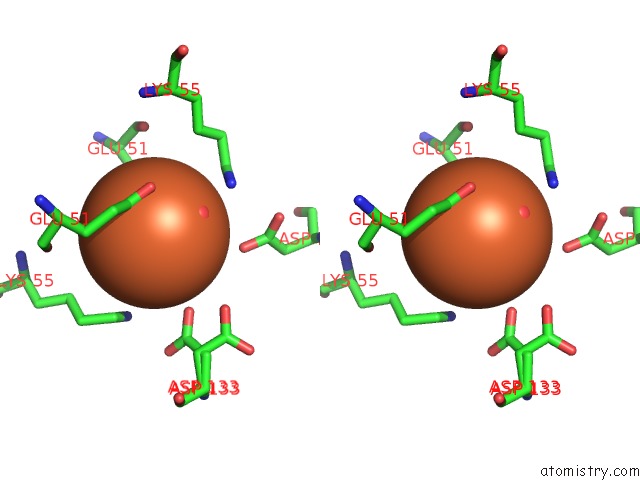
Stereo pair view

Mono view

Stereo pair view
A full contact list of Iron with other atoms in the Fe binding
site number 1 of Crystal Structure of Relp (SAS2) From Staphylococcus Aureus Bound to Pppgpp in the Post-Catalytic State within 5.0Å range:
|
Reference:
M.C.Manav,
J.Beljantseva,
M.S.Bojer,
T.Tenson,
H.Ingmer,
V.Hauryliuk,
D.E.Brodersen.
Structural Basis For (P)Ppgpp Synthesis By Thestaphylococcus Aureussmall Alarmone Synthetase Relp. J. Biol. Chem. V. 293 3254 2018.
ISSN: ESSN 1083-351X
PubMed: 29326162
DOI: 10.1074/JBC.RA117.001374
Page generated: Wed Aug 6 05:53:47 2025
ISSN: ESSN 1083-351X
PubMed: 29326162
DOI: 10.1074/JBC.RA117.001374
Last articles
Fe in 6I40Fe in 6I43
Fe in 6I3J
Fe in 6I3T
Fe in 6I3K
Fe in 6I36
Fe in 6I2Z
Fe in 6I2J
Fe in 6I0R
Fe in 6HWH