Iron »
PDB 6g71-6gl6 »
6gk5 »
Iron in PDB 6gk5: Crystal Structure of Cytochrome P450 CYP267B1 From Sorangium Cellulosum So CE56
Protein crystallography data
The structure of Crystal Structure of Cytochrome P450 CYP267B1 From Sorangium Cellulosum So CE56, PDB code: 6gk5
was solved by
I.K.Jozwik,
A.M.W.H.Thunnissen,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 41.00 / 1.60 |
| Space group | P 21 21 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 51.501, 66.390, 104.304, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 18.7 / 20.8 |
Iron Binding Sites:
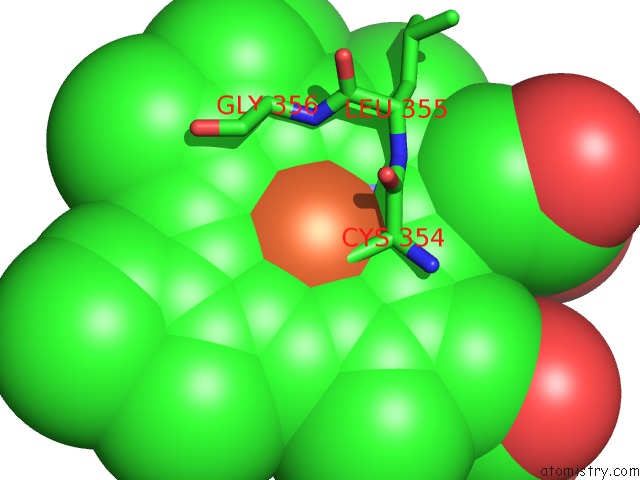
The binding sites of Iron atom in the Crystal Structure of Cytochrome P450 CYP267B1 From Sorangium Cellulosum So CE56
(pdb code 6gk5). This binding sites where shown within
5.0 Angstroms radius around Iron atom.
In total only one binding site of Iron was determined in the Crystal Structure of Cytochrome P450 CYP267B1 From Sorangium Cellulosum So CE56, PDB code: 6gk5:
In total only one binding site of Iron was determined in the Crystal Structure of Cytochrome P450 CYP267B1 From Sorangium Cellulosum So CE56, PDB code: 6gk5:
Iron binding site 1 out of 1 in 6gk5
Go back to
Iron binding site 1 out
of 1 in the Crystal Structure of Cytochrome P450 CYP267B1 From Sorangium Cellulosum So CE56
Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Iron with other atoms in the Fe binding
site number 1 of Crystal Structure of Cytochrome P450 CYP267B1 From Sorangium Cellulosum So CE56 within 5.0Å range:
|
Reference:
I.K.Jozwik,
M.Litzenburger,
Y.Khatri,
A.Schifrin,
M.Girhard,
V.Urlacher,
A.W.H.Thunnissen,
R.Bernhardt.
Structural Insights Into Oxidation of Medium-Chain Fatty Acids and Flavanone By Myxobacterial Cytochrome P450 CYP267B1. Biochem. J. V. 475 2801 2018.
ISSN: ESSN 1470-8728
PubMed: 30045877
DOI: 10.1042/BCJ20180402
Page generated: Wed Aug 6 06:54:09 2025
ISSN: ESSN 1470-8728
PubMed: 30045877
DOI: 10.1042/BCJ20180402
Last articles
Fe in 6KW7Fe in 6KY7
Fe in 6KQ1
Fe in 6KRF
Fe in 6KPS
Fe in 6KRC
Fe in 6KOE
Fe in 6KOC
Fe in 6KOF
Fe in 6KOB