Iron »
PDB 6r3w-6rr4 »
6rko »
Iron in PDB 6rko: Cryo-Em Structure of the E. Coli Cytochrome Bd-I Oxidase at 2.68 A Resolution
Enzymatic activity of Cryo-Em Structure of the E. Coli Cytochrome Bd-I Oxidase at 2.68 A Resolution
All present enzymatic activity of Cryo-Em Structure of the E. Coli Cytochrome Bd-I Oxidase at 2.68 A Resolution:
7.1.1.7;
7.1.1.7;
Iron Binding Sites:
The binding sites of Iron atom in the Cryo-Em Structure of the E. Coli Cytochrome Bd-I Oxidase at 2.68 A Resolution
(pdb code 6rko). This binding sites where shown within
5.0 Angstroms radius around Iron atom.
In total 3 binding sites of Iron where determined in the Cryo-Em Structure of the E. Coli Cytochrome Bd-I Oxidase at 2.68 A Resolution, PDB code: 6rko:
Jump to Iron binding site number: 1; 2; 3;
In total 3 binding sites of Iron where determined in the Cryo-Em Structure of the E. Coli Cytochrome Bd-I Oxidase at 2.68 A Resolution, PDB code: 6rko:
Jump to Iron binding site number: 1; 2; 3;
Iron binding site 1 out of 3 in 6rko
Go back to
Iron binding site 1 out
of 3 in the Cryo-Em Structure of the E. Coli Cytochrome Bd-I Oxidase at 2.68 A Resolution
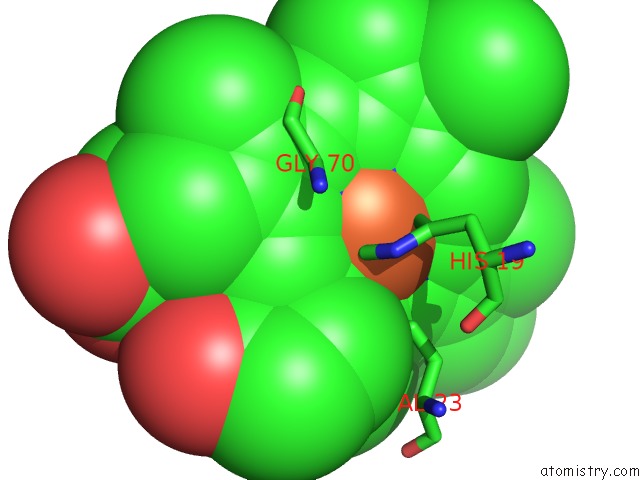
Mono view
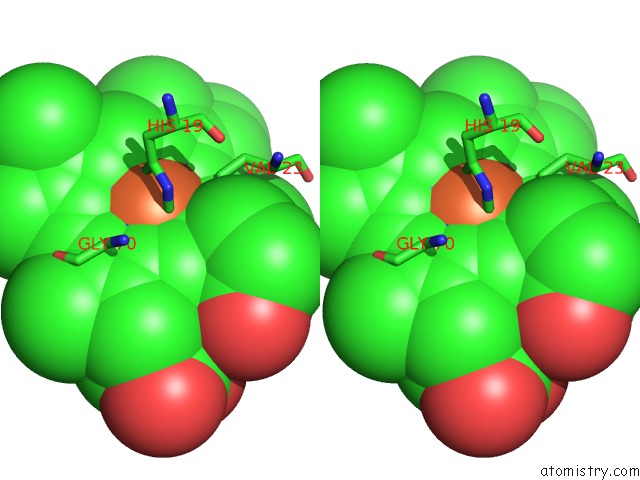
Stereo pair view

Mono view

Stereo pair view
A full contact list of Iron with other atoms in the Fe binding
site number 1 of Cryo-Em Structure of the E. Coli Cytochrome Bd-I Oxidase at 2.68 A Resolution within 5.0Å range:
|
Iron binding site 2 out of 3 in 6rko
Go back to
Iron binding site 2 out
of 3 in the Cryo-Em Structure of the E. Coli Cytochrome Bd-I Oxidase at 2.68 A Resolution
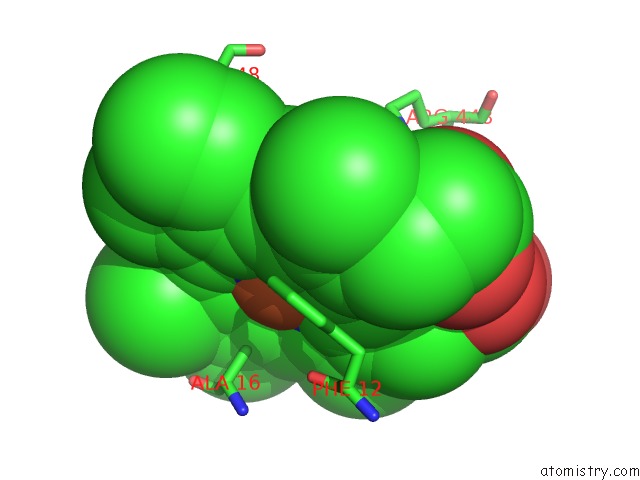
Mono view
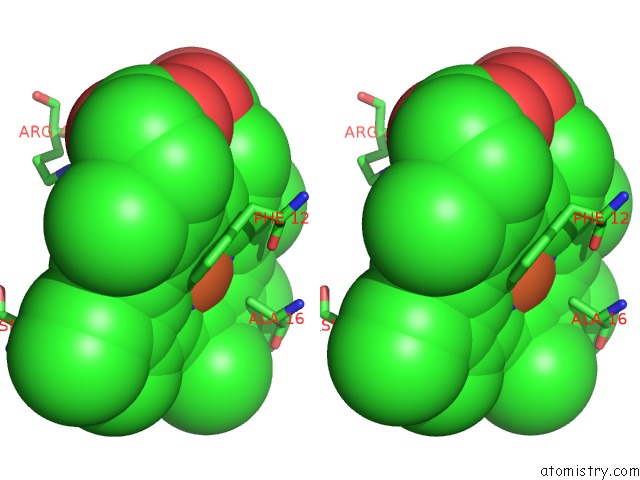
Stereo pair view

Mono view

Stereo pair view
A full contact list of Iron with other atoms in the Fe binding
site number 2 of Cryo-Em Structure of the E. Coli Cytochrome Bd-I Oxidase at 2.68 A Resolution within 5.0Å range:
|
Iron binding site 3 out of 3 in 6rko
Go back to
Iron binding site 3 out
of 3 in the Cryo-Em Structure of the E. Coli Cytochrome Bd-I Oxidase at 2.68 A Resolution
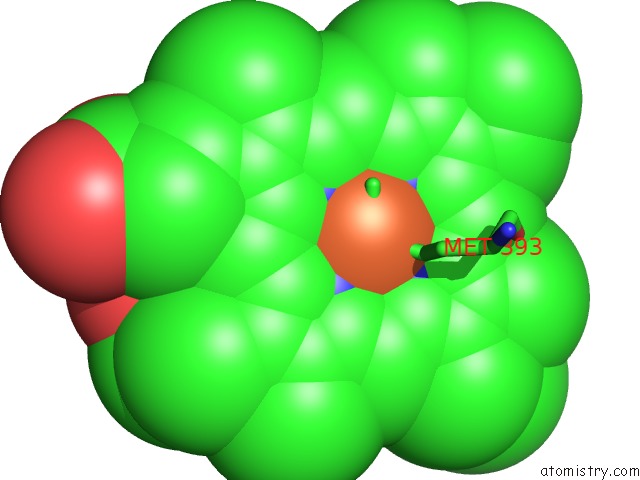
Mono view
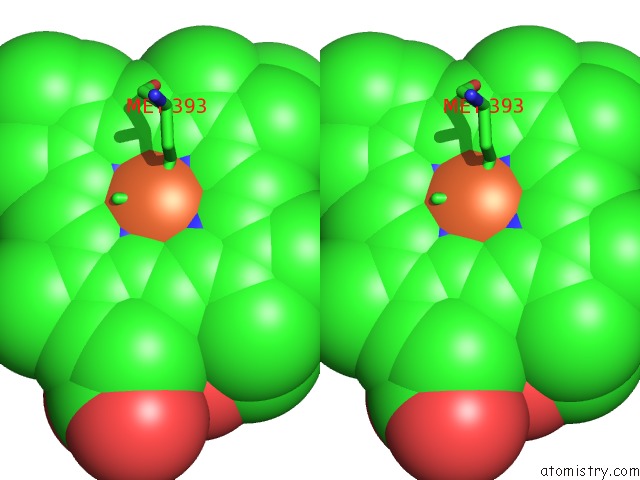
Stereo pair view

Mono view

Stereo pair view
A full contact list of Iron with other atoms in the Fe binding
site number 3 of Cryo-Em Structure of the E. Coli Cytochrome Bd-I Oxidase at 2.68 A Resolution within 5.0Å range:
|
Reference:
S.Safarian,
A.Hahn,
D.J.Mills,
M.Radloff,
M.L.Eisinger,
A.Nikolaev,
J.Meier-Credo,
F.Melin,
H.Miyoshi,
R.B.Gennis,
J.Sakamoto,
J.D.Langer,
P.Hellwig,
W.Kuhlbrandt,
H.Michel.
Active Site Rearrangement and Structural Divergence in Prokaryotic Respiratory Oxidases. Science V. 366 100 2019.
ISSN: ESSN 1095-9203
PubMed: 31604309
DOI: 10.1126/SCIENCE.AAY0967
Page generated: Wed Aug 6 13:06:39 2025
ISSN: ESSN 1095-9203
PubMed: 31604309
DOI: 10.1126/SCIENCE.AAY0967
Last articles
Fe in 7UV9Fe in 7UUR
Fe in 7UT9
Fe in 7UTE
Fe in 7UT8
Fe in 7UT6
Fe in 7UT7
Fe in 7USN
Fe in 7UEA
Fe in 7US8