Iron »
PDB 7qho-7r2s »
7qyz »
Iron in PDB 7qyz: Crystal Structure of A Dyp-Type Peroxidase 6E10 Variant From Pseudomonas Putida
Protein crystallography data
The structure of Crystal Structure of A Dyp-Type Peroxidase 6E10 Variant From Pseudomonas Putida, PDB code: 7qyz
was solved by
P.T.Borges,
D.Silva,
L.O.Martins,
C.Frazao,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 48.62 / 2.45 |
| Space group | P 2 3 |
| Cell size a, b, c (Å), α, β, γ (°) | 108.721, 108.721, 108.721, 90, 90, 90 |
| R / Rfree (%) | 22.2 / 25.5 |
Iron Binding Sites:
The binding sites of Iron atom in the Crystal Structure of A Dyp-Type Peroxidase 6E10 Variant From Pseudomonas Putida
(pdb code 7qyz). This binding sites where shown within
5.0 Angstroms radius around Iron atom.
In total only one binding site of Iron was determined in the Crystal Structure of A Dyp-Type Peroxidase 6E10 Variant From Pseudomonas Putida, PDB code: 7qyz:
In total only one binding site of Iron was determined in the Crystal Structure of A Dyp-Type Peroxidase 6E10 Variant From Pseudomonas Putida, PDB code: 7qyz:
Iron binding site 1 out of 1 in 7qyz
Go back to
Iron binding site 1 out
of 1 in the Crystal Structure of A Dyp-Type Peroxidase 6E10 Variant From Pseudomonas Putida
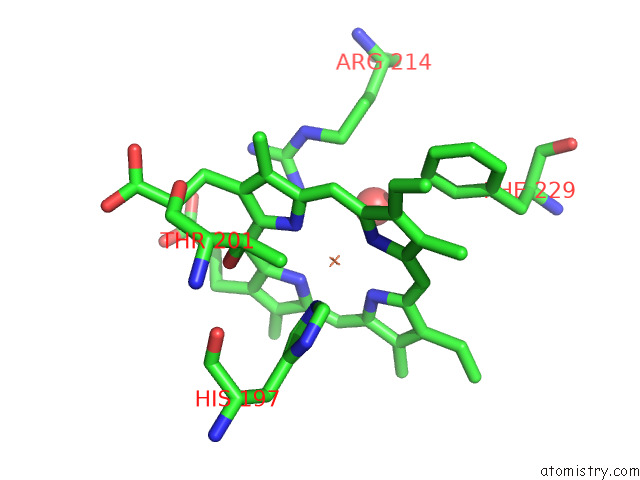
Mono view
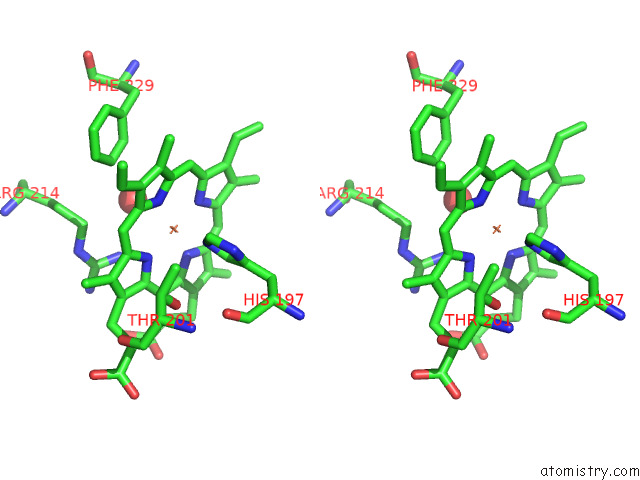
Stereo pair view

Mono view

Stereo pair view
A full contact list of Iron with other atoms in the Fe binding
site number 1 of Crystal Structure of A Dyp-Type Peroxidase 6E10 Variant From Pseudomonas Putida within 5.0Å range:
|
Reference:
P.T.Borges,
D.Silva,
T.F.D.Silva,
V.Brissos,
M.Canellas,
M.F.Lucas,
L.Masgrau,
E.P.Melo,
M.Machuqueiro,
C.Frazao,
L.O.Martins.
Unveiling Molecular Details Behind Improved Activity at Neutral to Alkaline pH of An Engineered Dyp-Type Peroxidase. Comput Struct Biotechnol J V. 20 3899 2022.
ISSN: ESSN 2001-0370
PubMed: 35950185
DOI: 10.1016/J.CSBJ.2022.07.032
Page generated: Thu Aug 7 04:10:59 2025
ISSN: ESSN 2001-0370
PubMed: 35950185
DOI: 10.1016/J.CSBJ.2022.07.032
Last articles
Fe in 7UTAFe in 7UWP
Fe in 7UVB
Fe in 7UVA
Fe in 7UV9
Fe in 7UUR
Fe in 7UT9
Fe in 7UTE
Fe in 7UT8
Fe in 7UT6